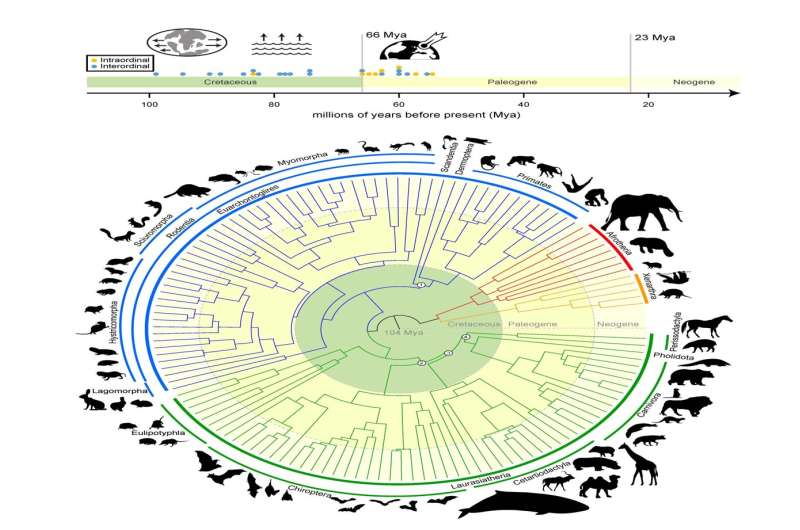
Foley’s efforts in the research study produced the world’s biggest mammalian phylogenetic tree to date. The “mammalian tree of life” draws up the development of mammals over more than 100 million years and is essential to the objectives of the Zoonomia Project. Credit: Texas A&M University
Research study led by a group of researchers from the Texas A&M School of Veterinary Medicine and Biomedical Sciences closes the heated clinical dispute relating to the history of mammal diversity as it associates with the termination of the non-avian dinosaurs. Their work offers a conclusive response to the evolutionary timeline of mammals throughout the last 100 million years.
The research study, released in Sciencebecomes part of a series of short articles launched by the Zoonomia Project, a consortium of researchers from around the world that is utilizing the biggest mammalian genomic dataset in history to identify the evolutionary history of the human genome in the context of mammalian evolutionary history. Their supreme objective is to much better recognize the hereditary basis for characteristics and illness in individuals and other types.
The Texas A&M University research study– led by Dr. William J. Murphy, a teacher in the Department of Veterinary Integrative Biosciences, and Dr. Nicole Foley, an associate research study researcher in Murphy’s laboratory– is rooted in phylogeny, a branch of biology that handles the evolutionary relationships and diversity of living and extinct organisms.
“The main argument has to do with whether placental mammals (mammals that establish within placentas) diverged prior to or after the Cretaceous-Paleogene (or K-Pg) termination occasion that erased the non-avian dinosaurs,” Foley shared. “By carrying out brand-new kinds of analyses just possible since of Zoonomia’s enormous scope, we respond to the concern of where and when mammals diversified and developed in relation to the K-Pg mass termination.”
The research study– which was performed with partners at the University of California, Davis; University of California, Riverside; and the American Museum of Natural History– concludes that mammals started diversifying prior to the K-Pg termination as the outcome of continental wandering, which triggered the Earth’s land masses to wander apart and return together over countless years. Another pulse of diversity happened instantly following the K-Pg termination of the dinosaurs, when mammals had more space, resources and stability.
This sped up rate of diversity caused the abundant variety of mammal family trees– such as predators, primates and hoofed animals– that share the Earth today.
Murphy and Foley’s research study was moneyed by the National Science Foundation and is one part of the Zoonomia Project led by Elinor Karlsson and Kerstin Lindblad-Toh, of the Broad Institute, which likewise compares mammal genomes to comprehend the basis of impressive phenotypes– the expression of particular genes such as brown vs. blue eyes– and the origins of illness.
Foley explained that the variety amongst placental mammals is displayed both in their physical qualities and in their amazing capabilities.
“Mammals today represent huge evolutionary variety– from the zooming flight of the small bumblebee bat to the sluggish slide of the massive Blue Whale as it swims through Earth’s large oceans. Several types have actually developed to echolocate, some produce venom, while others have actually developed cancer resistance and viral tolerance,” she stated.
“Being able to take a look at shared distinctions and resemblances throughout the mammalian types at a hereditary level can assist us determine the parts of the genome that are crucial to manage the expression of genes,” she continued. “Tweaking this genomic equipment in various types has actually caused the variety of qualities that we see throughout today’s living mammals.”
Murphy shared that Foley’s fixed phylogeny of mammals is important to the objectives of the Zoonomia Project, which intends to harness the power of relative genomics as a tool for human medication and biodiversity preservation.
“The Zoonomia Project is truly impactful since it’s the very first analysis to line up 241 varied mammalian genomes at one time and utilize that details to much better comprehend the human genome,” he described. “The significant inspiration for assembling this huge information set was to be able to compare all of these genomes to the human genome and after that identify which parts of the human genome have actually altered throughout mammalian evolutionary history.”
Figuring out which parts of genes can be controlled and which parts can not be altered without triggering damage to the gene’s function is very important for human medication. A current research study in Science Translational Medicine led by among Murphy and Foley’s coworkers, Texas A&M geneticist Dr. Scott Dindot, utilized the relative genomics approach to establish a molecular treatment for Angelman syndrome, a disastrous, uncommon neurogenetic condition that is set off by the loss of function of the maternal UBE3A gene in the brain.
Dindot’s group made the most of the very same procedures of evolutionary restriction recognized by the Zoonomia Project and used them to recognize an essential however formerly unidentified hereditary target that can be utilized to save the expression of UBE3A in human nerve cells.
Murphy stated broadening the capability to compare mammalian genomes by utilizing the biggest dataset in history will assist establish more remedies and treatments for other types’ disorders rooted in genes, consisting of felines and canines.
“For example, felines have actually physiological adjustments rooted in special anomalies that enable them to take in a specifically high-fat, high-protein diet plan that is incredibly unhealthy for people,” Murphy described. “One of the gorgeous elements of Zoonomia’s 241-species positioning is that we can choose any types (not simply human) as the recommendation and identify which parts of that types’ genome are totally free to alter and which ones can not endure modification. When it comes to felines, for instance, we might have the ability to assist determine hereditary adjustments in those types that might cause restorative targets for heart disease in individuals.”
Murphy and Foley’s phylogeny likewise played a critical function in a number of the subsequent documents that belong to the task.
“It’s trickle-down genomics,” Foley discussed. “One of the most pleasing things for me in working as part of the larger task was seeing the number of various research study jobs were improved by including our phylogeny in their analyses. This consists of research studies on preservation genomics of threatened types to those that took a look at the advancement of various intricate human qualities.”
Foley stated it was both significant and gratifying to definitively respond to the greatly discussed concern about the timing of mammal origins and to produce a broadened phylogeny that lays the structure for the next numerous generations of scientists.
“Going forward, this enormous genome positioning and its historic record of mammalian genome development will be the basis of whatever that everybody’s going to do when they’re asking relative concerns in mammals,” she stated. “That is quite cool.”
More info:
Nicole M. Foley et al, A genomic timescale for placental mammal development, Science (2023 ). DOI: 10.1126/ science.abl8189
Citation: Researchers utilize genomes of 241 types to redefine mammalian tree of life (2023, April 28) obtained 29 April 2023 from https://phys.org/news/2023-04-genomes-species-redefine-mammalian-tree.html
This file undergoes copyright. Apart from any reasonable dealing for the function of personal research study or research study, no part might be replicated without the composed authorization. The material is attended to details functions just.

